As part of research funded by the National Institutes of Health, the University of Denver Center for Orthopaedic Biomechanics has made available a repository of 3D models of the human lower extremities created from the Visible Human Female and Male imaging datasets.
Complete 3D musculoskeletal geometries were extracted from the National Libraries of Medicine Visible Human Female and Male cryosections. Muscle, bone, cartilage, ligament, and fat from the pelvis to the ankle were digitized and exported in shareable formats and made available for download. While a substantial amount of published work has been derived from the Visible Human Project, this is the first time a large number of musculoskeletal 3D geometries are being made available to the public that include both the male and female specimens. In total 260 geometries from the Visible Human Male and Female were extracted from the cryosections consisting of 76 muscles, 28 bones, 16 cartilages, 8 ligaments, and 2 fat geometries per subject. The library is available at multiple layers of processing and remarkably in a final form with no overlap between individual structures. This library is made available to motivate continued work in multi-scale, high-fidelity musculoskeletal modeling and promote reuse and continued development of the dataset.
Additional background can be found at https://ritchieschool.du.edu/research-innovation/biomedical-devices/center-orthopaedic-biomechanics and www.simtk.org/projects/3d-vh-geometry.
Support
This data was made possible by NIH grant U01 AR072989 with combined support from the National Institute for Arthritis, Musculoskeletal, and Skin Diseases (NIAMS), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and the National Institute of Child Health and Human Development (NICHD).
Methods
Courtesy of the U.S. National Library of Medicine, images were acquired for both the Visible Human Male (39M, 71 in., 90 kg, 27.8 BMI) and Female (59F, 62 in., 88 kg, 36 BMI). These images consist of digitized axial cryosection color photographs taken at 1 mm intervals in the VHM and 0.33 mm intervals in the VHF. The creation of the 3D models followed these steps:
- The cryosection and CT images were downloaded from the National Library of Medicine (https://www.nlm.nih.gov/research/visible/visible_human.html), reduced in size for manageability, and imported into software used for segmenting geometries from medical images (ScanIP v. S-2021.06, Simpleware, Synopsys, Mountain View, CA). Image volumes were cropped to the sternum. Offsets were created using original Cartesian coordinates of images from the original dataset, and manual/automatic alignment registration in non-aligning areas.
- Segmenting entailed manually selecting geometries in images using ScanIP. The researchers performing the segmentation referenced multiple anatomical sources, primarily Netter and Fleckenstein et al., and anatomical imaging applications at Radiopedia (www.radiopaedia.org) and AnatomyLearning (www.anatomylearning.com). Primal Pictures (Informa UK Limited, London) software was used as a reference for the Visible Human Male. Segmentation masks from Step 2 were exported from ScanIP as metaimage header (MHD) files to enable alteration or refinement of our digitization process by subsequent users. MHD files were included as the primary files containing the raw segmentation for use with most other commercial software, such as Mimics (Materialise, Belgium), ScanIP, and Amira (ThermoFisher Scientific, Waltham, MA).
- MHD files were combined in 3D Slicer (v. 4.11.20210226, www.slicer.org) to create nearly raw raster data (NRRD) files, the standard files used in 3D Slicer for segmentation. 3D slicer is a freely available software for reconstruction of three-dimensional geometry from medical imaging. NRRD files were included to allow for end users to quickly use 3DSlicer to view and edit segmentation in a freely available environment. NRRD masks were converted to TIFF stacks and binary labelmaps in .mat format for use in MATLAB (v. 2020b, Mathworks, Natick, MA). Muscle, bone, cartilage, ligament, and fat from the pelvis to the ankle were digitized and exported from ScanIP as raw 3D stereolithography (STL) objects without any post-processing. These raw 3D geometries may be used by others who wish to apply alternative means of post-processing than that described in steps 4 and 5.
- The raw STL geometries were then imported into MeshMixer (v. 3.5.474, AutoDesk, Rock Hill, SC) to reduce mesh sizes, remove sharp edges, and provide a smooth geometry without substantial alteration of the original geometry. Each geometry was remeshed to the following target edge lengths: muscle 1.5 mm, bone 1.0 mm, cartilage 0.75 mm, ligament 0.75 mm. In addition, segmentation masks were generated from the processed STL files for those who wish to modify the resampled and smoothed geometries in 3D Slicer.
- Overclosures between the resampled-smoothed 3D geometries were removed using custom MATLAB code utilizing radial basis functions. All surface overclosures were removed to achieve a minimum gap distance of 0.05 mm between all geometries. Pairs of geometries that were overclosed with one of the geometries as bone, were set to remove overclosures by only deforming nodes on the mating geometry. In all other cases, overclosures were removed by deforming both geometries equally. The final geometries were exported as 3D STL objects. In addition, segmentation masks were generated for those who wish to modify the final geometries in 3D Slicer.
Data Records
In total 260 geometries from the Visible Human Male and Female were extracted. The skeletal components consist of the pelvis through the feet (Table 1); the ligament and cartilage components consist of hip, knee, and ankle cartilages and knee ligaments (Table 2); muscle components consist of 76 separate muscles from the Iliacus proximally to the Flexor Digitorum distally (Table 3), and two fat components consisting of the intramuscular fat and fascia, and the outer fat, dermis, and epidermis (Table 4).
Table 1: The 15 skeletal components available for download in both the female and male subjects. Lower limb bones are unique on the right and left sides of the subjects (resulting in 28 unique bones per subject).
| Bones of the Lower Extremities | ||
| Coccyx Sacrum Pelvis (Ischium + Illium + Pubis) x 2 |
Femur x 2 Patella x 2 Tibia x 2 Fibula x 2 |
Talus x 2 Calcaneus x 2 Navicular x 2 Cuboid x 2 Lateral Cuneiform x 2 Medial Cuneiform x 2 Intermediate Cuneiform x 2 Phalanges (Tarsals + Metatarsals + Phalanges) x 2 |
Table 2: The 12 identified ligaments and cartilage components available for download in both the female and male subjects. Ligaments and cartilage are unique on the right and left sides of the subjects (resulting in 24 unique tissues per subject).
| Knee Ligaments | Articular Cartilages |
| Anterior Cruciate x 2 Posterior Cruciate x 2 Medial Collateral x 2 Lateral Collateral x 2 |
Femoroacetabular (Femoral Head + Pelvis Acetabulum) x 2 Tibiofemoral (Distal Femur + Lateral Tibia + Medial Tibia) x 2 Patellofemoral (Distal Femur + Patellar) x 2 Tibiotalar (Distal Tibia + Proximal Talus) x 2 |
Table 3: The 38 identified muscle components available for download in both the female and male subjects. Muscle components are unique on the right and left sides of the subjects (resulting in 76 unique muscle geometries per subject).
| Muscles of the Lower Extremities | ||
| Adductor Brevis x 2 Adductor Longus x 2 Adductor Magnus x 2 Biceps Femoris Long x 2 Biceps Femoris Short x 2 Extensor Digitorum Longus x 2 Extensor Hallucis Longus x 2 Flexor Digitorum Longus x 2 Flexor Hallucis Longus x 2 Gastrocnemius Lateral x 2 Gastrocnemius Medial x 2 Gluteus Maximus x 2 Gluteus Medius x 2 |
Gluteus Minimus x 2 Gracilis x 2 lliacus x 2 lnferior Gemellus x 2 Obturator Externus x 2 Obturator lnternus x 2 Pectineus x 2 Peroneus Longus x 2 Piriformis x 2 Plantaris x 2 Popliteus x 2 Psoas Major x 2 Quadratus Femoris x 2 |
Rectus Femoris x 2 Sartorius x 2 Semimembranosus x 2 Semitendonosus x 2 Soleus x 2 Superior Gemellus x 2 Tensor Fasciae Latae x 2 Tibialis Anterior x 2 Tibialis Posterior x 2 Vastus lntermedius x 2 Vastus Lateralis x 2 Vastus Medialis x 2 |
Table 4: The 2 identified fat components available for download in both the female and male subjects. Fat components are unique on the right and left sides of the subjects but contained in single files (resulting in 2 unique fat geometries per subject).
| Fat and Fasciae Components | |
| Intermuscular fat and fasciae | Outer fat (epidermis + dermis + fat) |
License
Creative Commons License
This work is licensed under a Creative Commons Attribution 4.0 International License
Liability Agreement
The Data is provided “as is” with no express or implied warranty or guarantee. The University of Denver and the Center for Orthopaedic Biomechanics do not accept any liability or provide any guarantee in connection with uses of the Data, including but not limited to, fitness for a particular purpose and noninfringement. The University of Denver and the Center for Orthopaedic Biomechanics are not liable for direct or indirect losses or damage, of any kind, which may arise through the use of this data.
Citations
T. E. Andreassen, D. R. Hume, L. D. Hamilton, K. E. Walker, S. E. Higinbotham, and K. B. Shelburne, “Three Dimensional Lower Extremity Musculoskeletal Geometry of the Visible Human Female and Male,” Sci. Data, vol. 10, no. 1, p. 34, Jan. 2023, doi: 10.1038/s41597-022-01905-2.
T. E. Andreassen, D. R. Hume, L. D. Hamilton, S. E. Higinbotham, and K. B. Shelburne, “An Automated Process for 2D and 3D Finite Element Overclosure and Gap Adjustment using Radial Basis Function Networks,” arXiv Comput. Sci., Sep. 2022, doi: https://doi.org/10.48550/arXiv.2209.06948
DOI for this page: https://doi.org/10.56902/COB.vh.2022.0
Adding new geometries is encouraged! New geometries can be added to the dataset by contacting the authors (kevin.shelburne@du.edu and thor.andreassen@du.edu ). The authors will check new or revised content for accuracy and completeness and update the folders with credit of the contributors highlighted on this website.
Additional Contributors
Daniella Duran, The University of Denver
Victoria Volk, Clare Fitzpatrick, and Colton Babcock, Boise State University
Jenelys Cox, The University of Denver
-
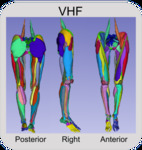
Visible Human Female
Center for Orthopaedic Biomechanics, Thor E. Andreassen, Donald R. Hume, Landon D. Hamilton, Karen E. Walker, Sean E. Higinbotham, and Kevin B. Shelburne
Objects Available for Download
- Aligned Cryosection Images: moving proximal to distal in the Visible Human sequences of cryosection images, there are some offsets in the transverse plane that require correction before beginning segmentation. The Visible Human Female images contain some challenging offsets. As correction is a time-consuming process, we have made the corrected images available for download. The corrected scans are available in DICOM, TIFF, .mat, and MHD file formats.
- Aligned and Rescaled CT Images: the Visible Human CT images are useful for segmentation of tissues that are not as clear in the cryosection images. However, the CT images are not precisely aligned with the cryosection images. We have made available the CT images aligned to the cryosection images also with offsets corrected. CT scans are full scans going from the head-to-toes. The corrected scans are available in DICOM, TIFF, and MHD file formats.
- Original Segmentation Masks: the 3D models were created using ScanIP by the construction of 3D objects from a series of outlines, or masks, of each object. This was a manual process often requiring subjective decisions when the clarity of the images made the detection of tissue borders challenging. Therefore, we have provided the segmentation masks in 3D Slicer for those that wish to check or alter the masks for creation of unique models. The raw segmentation masks are available in 3D Slicer as NRRD files. Additionally, the segmentations are available as binary label maps in MHD, TIFF, and .mat file formats.
- Raw 3D Models: the raw 3D models created from ScanIP are provided in STL format in the default edge length of ScanIP, approximately 0.33 mm edge lengths. Providing the raw STL models enables others to apply their own preferred means of post-processing of the objects (e.g., smoothing or lofting) starting from their original state.
- Smoothed 3D Models: the smoothed and resampled STLs from MeshMixer are provided. These geometries are free of issues resulting from segmentation and each geometry was remeshed to the following target edge lengths: muscle 1.5 mm, bone 1.0 mm, cartilage 0.75 mm, ligament 0.75 mm.
- Segmentation Masks of Smoothed Models: segmentation masks were created from the smoothed and resampled 3D models to enable transverse inspection of the final product in segmentation software 3D Slicer.
- Final 3D Models: the goal of the project was to provide 3D models of the tissues that could be used in applications without further processing. The final 3D models were visually inspected to ensure no existing sharp edges and then checked and corrected for any overlap or overclosure. If an overclosure was present, it was removed to provide a gap distance of 0.05 mm using a unique radial basis function-based MATLAB code that is publicly available at: https://github.com/thor-andreassen/femors and described in Andreassen et al. (in review) 2022.
- Segmentation Masks of Final Models: segmentation masks were created from the final 3D models to enable transverse inspection of the final product in segmentation software 3D Slicer.
- Comparison Metadata: includes tables of the initial overclosure amounts tissue geometries as well as comparisons between tissue volumes before and after smoothing and overclosure correction. Comparisons are also made between tissue volumes on the left and right side of the body.
Folders for Download
The complete dataset is very large. For this reason, the datasets have been split into manageable folders for download:
- Aligned Cryosection DICOM
Zip size: 462.8 MB
Extracted size: 485.0 MB - Aligned CT DICOM
Zip size: 302.0 MB
Extracted size: 316.0 MB - Aligned Scan Images (.mat, .tif, .ctbl)
Zip size: 1472.6 MB
Extracted size: 1540.0 MB - Original segmentation masks and Aligned scans (.mhd) (Right, Left, or Combined)
Zip size: 1318.1 MB
Extracted size: 129,000.0 MB - Original segmentation label maps (.mat, .tif)
Zip size: 2081.3 MB
Extracted size: 2180.0 MB - Original segmentation masks and Aligned scans (3D Slicer) (Right, Left, or Combined, Combined with CT)
Zip size: 2616.3 MB
Extracted size: 2740.0 MB - Smoothed Segmentation masks and Aligned Scans (3D Slicer) (Right, Left, or Combined)
Zip size: 2073.7 MB
Extracted size: 2180.0 MB - Final Segmentation masks and Aligned Scans (3D Slicer) (Right, Left, or Combined)
Zip size: 2076.0 MB
Extracted size: 2180.0 MB - Original 3D STL models (Right, Left, or Combined)
Zip size: 506.2 MB
Extracted size: 5660.0 MB - Smoothed 3D STL models (Right or Left)
Zip size: 173.0 MB
Extracted size: 601.0 MB - Final 3D STL models (Right or Left)
Zip size: 87.8 MB
Extracted size: 117.0 MB - Metadata
-
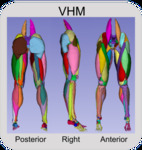
Visible Human Male
Center for Orthopaedic Biomechanics, Thor E. Andreassen, Donald R. Hume, Landon D. Hamilton, Karen E. Walker, Sean E. Higinbotham, and Kevin B. Shelburne
Objects Available for Download
- Aligned Cryosection Images: moving proximal to distal in the Visible Human sequences of cryosection images, there are some offsets in the transverse plane that require correction before beginning segmentation. The Visible Human Female images contain some challenging offsets. As correction is a time-consuming process, we have made the corrected images available for download. The corrected scans are available in DICOM, TIFF, .mat, and MHD file formats.
- Aligned and Rescaled CT Images: the Visible Human CT images are useful for segmentation of tissues that are not as clear in the cryosection images. However, the CT images are not precisely aligned with the cryosection images. We have made available the CT images aligned to the cryosection images also with offsets corrected. CT scans are full scans going from the head-to-toes. The corrected scans are available in DICOM, TIFF, and MHD file formats.
- Original Segmentation Masks: the 3D models were created using ScanIP by the construction of 3D objects from a series of outlines, or masks, of each object. This was a manual process often requiring subjective decisions when the clarity of the images made the detection of tissue borders challenging. Therefore, we have provided the segmentation masks in 3D Slicer for those that wish to check or alter the masks for creation of unique models. The raw segmentation masks are available in 3D Slicer as NRRD files. Additionally, the segmentations are available as binary label maps in MHD, TIFF, and .mat file formats.
- Raw 3D Models: the raw 3D models created from ScanIP are provided in STL format in the default edge length of ScanIP, approximately 0.33 mm edge lengths. Providing the raw STL models enables others to apply their own preferred means of post-processing of the objects (e.g., smoothing or lofting) starting from their original state.
- Smoothed 3D Models: the smoothed and resampled STLs from MeshMixer are provided. These geometries are free of issues resulting from segmentation and each geometry was remeshed to the following target edge lengths: muscle 1.5 mm, bone 1.0 mm, cartilage 0.75 mm, ligament 0.75 mm.
- Segmentation Masks of Smoothed Models: segmentation masks were created from the smoothed and resampled 3D models to enable transverse inspection of the final product in segmentation software 3D Slicer.
- Final 3D Models: the goal of the project was to provide 3D models of the tissues that could be used in applications without further processing. The final 3D models were visually inspected to ensure no existing sharp edges and then checked and corrected for any overlap or overclosure. If an overclosure was present, it was removed to provide a gap distance of 0.05 mm using a unique radial basis function-based MATLAB code that is publicly available at: https://github.com/thor-andreassen/femors and described in Andreassen et al. (in review) 2022.
- Segmentation Masks of Final Models: segmentation masks were created from the final 3D models to enable transverse inspection of the final product in segmentation software 3D Slicer.
- Comparison Metadata: includes tables of the initial overclosure amounts tissue geometries as well as comparisons between tissue volumes before and after smoothing and overclosure correction. Comparisons are also made between tissue volumes on the left and right side of the body.
Folders for Download
The complete dataset is very large. For this reason, the datasets have been split into manageable folders for download:
- Aligned Cryosection DICOM
Zip size: 607.0 MB
Extracted size: 578.9 MB - Aligned CT DICOM
Zip size: 291.0 MB
Extracted size: 278.4 MB - Aligned Scan Images (.mat, .tif, .ctbl)
Zip size: 1988.8 MB
Extracted size: 1950.0 MB - Original segmentation masks and Aligned scans (.mhd) (Right, Left, or Combined)
Zip size: 1527.0 MB
Extracted size: 191,000.0 MB - Original segmentation label maps (.mat, .tif)
Zip size: 3883.2 MB
Extracted size: 4070.0 MB - Original segmentation masks and Aligned scans (3D Slicer) (Right, Left, or Combined, Combined with CT)
Zip size: 3260.3 MB
Extracted size: 3410.0 MB - Smoothed Segmentation masks and Aligned Scans (3D Slicer) (Right, Left, or Combined)
Zip size: 2773.1 MB
Extracted size: 2910.0 MB - Final Segmentation masks and Aligned Scans (3D Slicer) (Right, Left, or Combined)
Zip size: 2773.0 MB
Extracted size: 2910.0 MB - Original 3D STL models (Right, Left, or Combined)
Zip size: 1296.5 MB
Extracted size: 8380.0 MB - Smoothed 3D STL models (Right or Left)
Zip size: 244.0 MB
Extracted size: 793.0 MB - Final 3D STL models (Right or Left)
Zip size: 139.0 MB
Extracted size: 238.0 MB - Metadata
